SARS-CoV-2 genome analysis
I took a class called “Bio-Data Structures” in my second year. Don’t pay attention to the name, the class had nothing to do with data structures. It was based on the famous Compeau and Pevzner book, and was aimed to introduce programming principles and to teach how to translate biological problems into well-defined computational ones. In the final project, we were asked to analyze SARS-CoV-2 genomes from April 19, 2021 to June 28, 2021. Even though I ended up doing more computational chemistry/biophysics things rather than computational omics, I quite enjoyed the course and it ignited my interest in computational biology. Below is a modified version of my final report.
The dataset was in .fasta format, which is basically just text files with the genomic sequences and some metadata. The analysis pipeline was split into three main steps:
- Run a multiple sequence alignment (MSA) against a reference spike RNA sequence.
- Scan the aligned sequences for mutations and dump them into
.csvfiles. - Plot the mutation frequencies to see how they evolve over time.
Multiple Sequence Alignment (MSA)
Let’s start with the alignment part. The algorithm we used is heuristic: instead of aligning all sequences at once (which is technically the “optimal” way but computationally insane), it builds up a profile from previously aligned sequences and matches new ones against it. That means it’s not perfectly accurate, since the algorithm doesn’t know what future sequences will look like. But in this case, the genomes are all from the same virus, collected within just a couple of months, so the accuracy loss is predicted (and assumed) to be acceptable.
For context: the heuristic method is O(n²) in both time and space. The “true” optimal alignment would blow up to O(nᴺ) because it tries to align all N sequences in one giant multidimensional graph. One can reduce the space complexity to O(n) using divide-and-conquer tricks when facing memory limitations, but then you double the runtime.
The scoring system we used was slightly tweaked compared to the defaults:
- match = +1
- mismatch = 0
- indel (insertion/deletion) = −5
The penalties are a bit lower than widely used numbers as we know the genomic sequences are of the same organism, and the organism studied in this project is an RNA virus with a tendency to quickly mutate, so we should allow a small number of mismatches and indels by design and avoid overfitted sequences.
Mutation Detection
Once the sequences are aligned, we start looking for mismatches and count how often they show up. Each mismatch/indel is considered one single-nucleotide polymorphism. Such simple approach helps us to find these mutations very easily, without major limitations. The main limitation of this approach is the inability to find more complex limitations, i.e., mutations other than single-nucleotide mutations. So, when this simple approach is used, subsequent manual analyses is needed to find more complex mutations and extract more information from the genomic sequences.
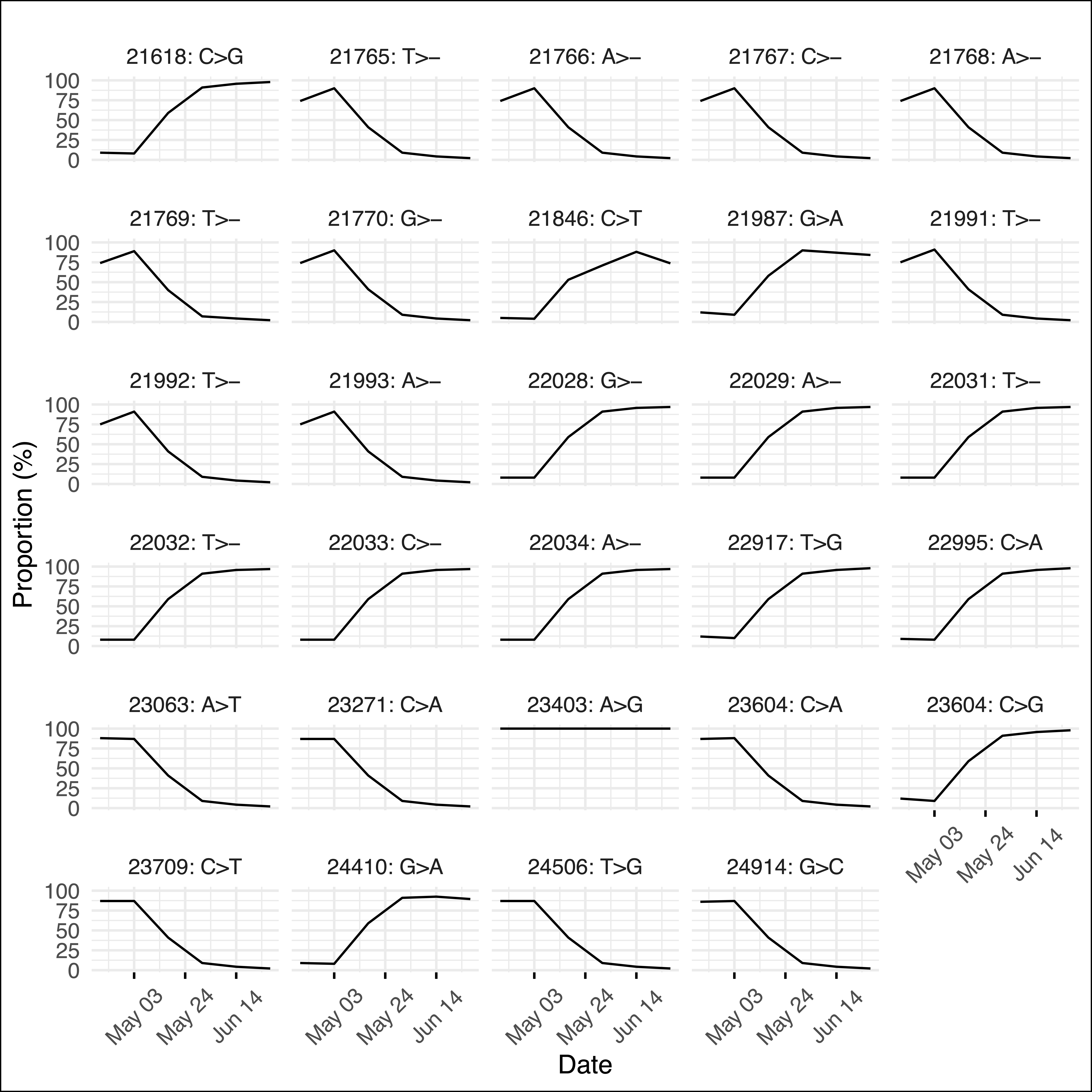
To make the mutation data more biologically meaningful, we adjust the nucleotide positions. For example, the most frequent mutation in our dataset was an A→G change at position 1844. When compared to the reference SARS-CoV-2 genome, that actually maps to position 23403, which sits right inside the S gene (the one coding for the spike protein). Once we shifted the numbering baseline, everything lined up correctly with known mutations.
Here are the most common single-nucleotide mutations we found:
| Mutation index | Nucleotide(s)’ position | Nucleotide(s) change | Cumulative frequency in assignment dataset | Cumulative frequency in open-source dataset |
|---|---|---|---|---|
| 1 | 21618 | C>G | 59.4 | 77.8 |
| 2 | 21765 | TACATG>- | 37.4 | 21.1 |
| 3 | 21846 | C>T | 48.5 | 43.4 |
| 4 | 21987 | G>A | 56.1 | 47.6 |
| 5 | 21991 | TTA>- | 37.8 | 21.4 |
| 6* | 22028 | GATTCA>- | 59.0 | - |
| 7 | 22029 | AGTTCA>- | 59.0 | 74.9 |
| 8 | 22917 | T>G | 60.2 | 77.0 |
| 9 | 22995 | C>A | 59.4 | 76.7 |
| 10 | 23063 | A>T | 39.3 | 21.0 |
| 11 | 23271 | C>A | 39.1 | 21.2 |
| 12 | 23403 | A>G | 100 | 99.98 |
| 13 | 23604 | C>G | 60.0 | 78.0 |
| 14 | 23604 | C>A | 39.3 | 21.5 |
| 15 | 23709 | C>T | 39.1 | 21.2 |
| 16 | 24410 | G>A | 57.5 | 76.7 |
| 17 | 24506 | T>G | 39.1 | 21.2 |
| 18 | 24914 | G>C | 38.9 | 21.2 |
*excluded from the graph as the mutation is not found in open-source dataset
Interestingly, while the frequencies in our small dataset didn’t exactly match the open-source dataset, the trends were consistent. If you plot them, the correlation is basically a straight line with slope ~1, intercept 0. The small discrepancies are just sampling bias: our 100 genomes weren’t fully “representative,” while the open-source dataset is much larger.

Multiple-Nucleotide Mutations
Now, we can see that there are 3 or 6 indels show up right after the other. For example, the sets of mutations at positions 21765–21770, 21991–21993, and 22029–22034 clearly move together. We can combine these the single-nucleotide mutations into a multiple-nucleotide mutation, and this combination is corroborated by statistical and biological evidence:
- In the assignment dataset, the frequency plots of the single-nucleotide changes are identical in both frequency and time. Thus, there is a strong chance these changes are connected.
- In the open-source dataset, the frequency of only one or few of these single-nucleotide changes is much smaller than the frequency of the multiple-nucleotide mutation.
- These nucleotide changes occur for 3 or 6 nucleotides, which is equal to the length of 1 and 2 codons, respectively. So, when 3 or 6 nucleotides are deleted, a whole codon or 2 codons are dropped, which indicates an amino acid deletion mutation.
From Nucleotides to Amino Acids
Once you know the nucleotide positions, it’s straightforward to map them to amino acids using a codon table. That lets you check which changes are synonymous (don’t affect the protein sequence) versus non-synonymous (do change the protein sequence).
| Mutation index | Nucleotide(s)’ position | Nucleotide(s) change | Amino acid change |
|---|---|---|---|
| 1 | 21618 | C>G | T19R |
| 2 | 21765 | TACATG>- | HV 69–70 deletion |
| 3 | 21846 | C>T | T95I |
| 4 | 21987 | G>A | G142D |
| 5 | 21991 | TTA>- | Y144 deletion |
| 6* | 22028 | GATTCA>- | - |
| 7 | 22029 | AGTTCA>- | EF 156-157 deletion |
| 8 | 22917 | T>G | L452R |
| 9 | 22995 | C>A | T478K |
| 10 | 23063 | A>T | N501Y |
| 11 | 23271 | C>A | A570D |
| 12 | 23403 | A>G | D614G |
| 13 | 23604 | C>G | P681H |
| 14 | 23604 | C>A | P681R |
| 15 | 23709 | C>T | T716I |
| 16 | 24410 | G>A | D950N |
| 17 | 24506 | T>G | S982A |
| 18 | 24914 | G>C | D1118H |
This is where things get interesting, and I think this was the point of the whole project. Some of these mutations we found are known “defining mutations” of major variants:
- Delta (21A/S:478K): mutations 1, 4, 7, 8, 9, 12, 14, 16
- Alpha (S.501Y.V1): mutations 2, 5, 10, 11, 12, 13, 15, 17, 18
If you track their frequencies over time, you can actually see the variant takeover happening. Before May 3, Alpha mutations dominate (~75–80%) and Delta is still rare (~10%). After May 3, Alpha drops off and Delta rapidly rises. Mutation 12 is present in both, so it stays at 100%. This matches real-world data almost perfectly: by June 4, 2021, the Delta variant had officially become dominant in the UK.
Functional Impact
Using this data, we can also determine which mutations are more likely to be responsible for increased pathogenicity. One of such cases is the case of P681R and P681H mutations of the Delta and Alpha strands, respectively. It has been determined that the P681 mutation is key to the increased pathogenicity of the Delta variant. The reason behind the effectiveness of this mutation is the cleavage of S1 and S2 subunits which is predicted to improve cell-surface-mediated virus entry. The alpha variant also exhibits enhanced S1/S2 cleavage thanks to its P681H mutation, however, the P681R mutation is much more effective than the P681H mutation.